
The human genome contains nearly 3 billion bases, yet only about 1.7%, around 180,000 coding regions make up the exome. Remarkably, about 85% of disease-causing mutations are found within this small fraction. By focusing on the exome, whole exome sequencing (WES) delivers a powerful, cost-effective approach to uncover clinically relevant variants with far greater efficiency than whole genome sequencing. Whole-exome sequencing (WES) is a powerful next-generation sequencing (NGS) approach that decodes the protein-coding regions of the genome, the area's most responsible for disease, making it a widely adopted tool in clinical and research settings.
Whole-exome sequencing, powered by exome enrichment, is an efficient and powerful tool to uncover genetic variants that shape heritable traits ranging from disease-causing mutations to natural variations making it invaluable for applications in population genetics, cancer research, genetic disease studies, and even crop and livestock improvement. Conventional exome sequencing panels often require a trade-off between comprehensive genomic coverage and assay performance, leading to uneven coverage, higher duplication rates, ultimately impacting the sensitivity and reliability of variant detection.
Developed with a deep understanding of both clinical and research needs, G2M’s Whole exome sequencing assay is designed with exceptional uniformity and high on-target efficiency with the panel content aligned with the latest curated genomic data for enhanced clinical relevance. The panel encompasses ~21,500 genes catering to various hereditary conditions and germline cancers.
*Hybridization time may vary based on panel size
High Confidence Gene Annotation Across Trusted Databases
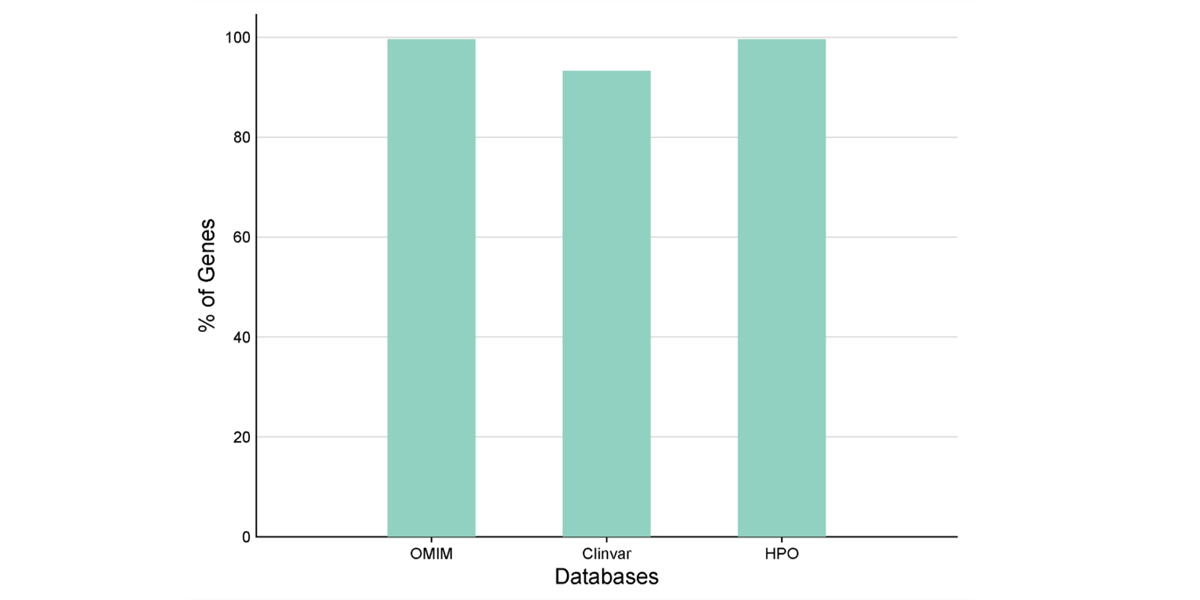
The chart illustrates the high percentage of genes mapped to major clinical and phenotype databases - OMIM, ClinVar, and HPO ensuring robust integration of genetic information. Nearly 100% coverage in OMIM and HPO, along with over 90% in ClinVar, highlights the robustness of these databases for accurate gene interpretation and evidence based clinical insights.
Optimized Coverage Delivering Accuracy Across Samples
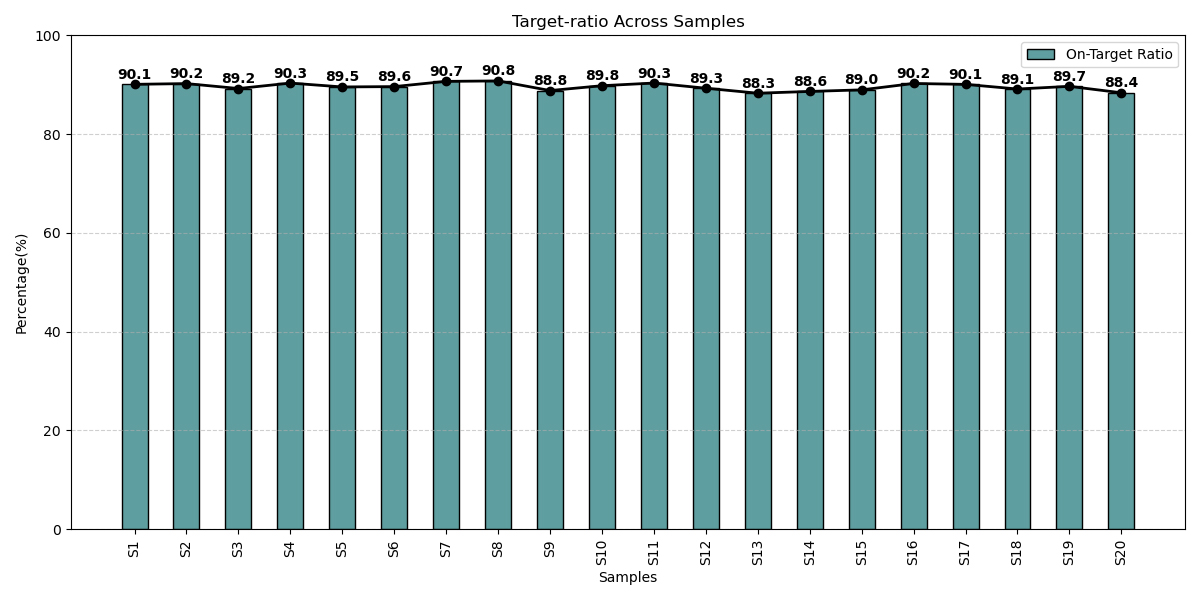
On-target ratios across patient samples consistently exceeded over 85%, highlighting the panel’s optimized design, efficient probe capture, and robust sequencing performance for reliable genomic profiling.
Exceptional Coverage Uniformity Validated by Low Fold 80 Metric
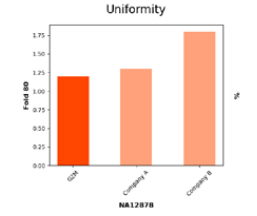
Fold 80 base penalty measures coverage uniformity, the lower the value, the less over-sequencing required for reliable results. G2M achieves a penalty of < 1.25, reflecting exceptional capture design and hybridization efficiency compared to competitors, ensuring cost-effective, high-quality sequencing.
Efficient Whole Exome Sequencing with Ultra-Low Depth Coverage

The bar chart compares target coverage at 1X for sample NA12878 across three panels: G2M, Company A, and Company B. G2M demonstrates near complete coverage (~100%), outperforming competitors and ensuring reliable sequencing with minimal gaps critical for accurate variant detection and high-quality results.
| Commercial Name | Cat No. | Pack Size | Platform |
|---|---|---|---|
| Clinical Exome Sequencing Expanded (WES) NGS Test kit | G710008-1 | 24 T | Illumina |
| G710008-2 | 96 T | Illumina | |
| G710008-3 | 96 T - EZY | Illumina - EZY | |
| Clinical Exome Sequencing Expanded (WES) NGS Test kit | G710008-4 | 24 T | MGI |
| G710008-5 | 96 T | MGI | |
| G710008-6 | 96 T - EZY | MGI - EZY | |
| Clinical Exome Sequencing Expanded (WES) NGS Test kit | G710008-7 | 24 T | Aviti |
| G710008-8 | 96 T | Aviti | |
| G710008-9 | 96 T - EZY | Aviti - EZY | |
| Clinical Exome Sequencing Expanded (WES) NGS Test kit | G710008-10 | 24 T | Thermo |
| G710008-11 | 96 T | Thermo | |
| G710008-12 | 96 T – EZY | Thermo – EZY |
Since its inception in 2016, Genes2me has been constantly striving towards setting a benchmark in the diagnostics space by introducing premium quality (Made in India) diagnostic kits which are CE-IVD, ISO-13485:2016, and ISO 9001:2015 certified, assuring our clients of unparalleled quality and compliance with international standards.
© 2025 Genes2me. All rights reserved.